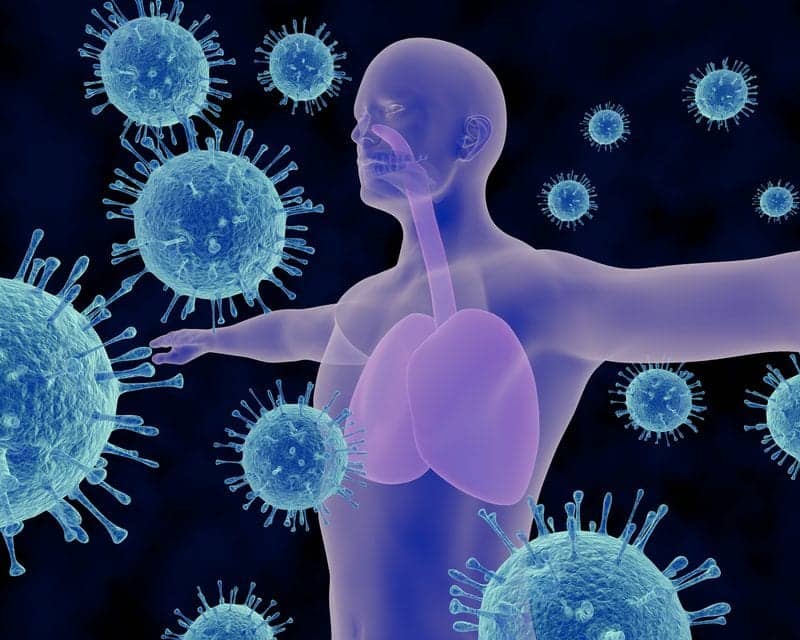
British researchers have applied population genetics to the vexing problem of Mycobacterium abscesses, a dangerous pathogen that is spreading through the cystic fibrosis community.
“We obtained maximum-likelihood phylogenetic trees demonstrating separation of M. abscessus into three clearly divergent subspecies (M a. abscessus, M. a bollettii, and M. a. massiliense), challenging recent reclassifications of M. abscessus into only two subspecies,” the researchers wrote.
Within each subspecies, Floto et al identified large genetic differences between isolates, but they also found multiple clades of near-identical isolates from geographically diverse locations — “suggesting widespread transmission of circulating clones within the global cystic fibrosis patient community.”
Subspecies phylogeny analysis, excluding clusters found at only one cystic fibrosis center, revealed that most patients (74%) were infected with clustered, rather than unclustered, isolates, mainly from M. a abscessus cluster 1 and 2 and massiliense cluster 1.
“The median bunch lengths of almost all clusters found in two or more cystic fibrosis centers was less than 20 single-nucleotide polymorphisms (SNPs), indicating a high frequency of identical isolates infecting geographically separate individuals,” the researchers wrote.
Further analysis strongly suggested that M. abscessus spreads though fomite and airborne means.
“Given the widespread implementation of individual and cohort segregation of patients in cystic fibrosis centers in Europe, the United States, and Australia, which have led to falling levels of MRSA, Burkholderia, and transmissible Pseudomonas infections, we believe that the likely mechanism of local spread of M. abscessus is via fomite spread or potentially through the generation of long-lived infectious aerosols.”
Get the full story at www.medpagetoday.com