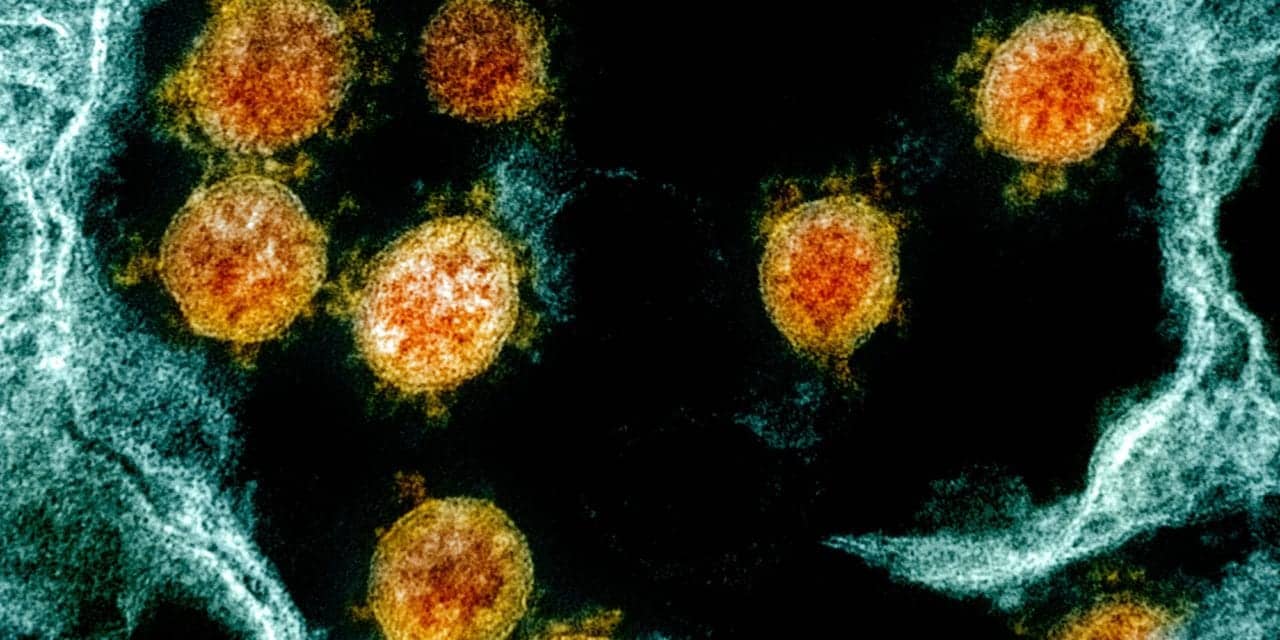
The US FDA awarded a new research contract to the Stanford University School of Medicine to perform an in-depth analysis of tissue samples to learn more about how SARS-CoV-2—the virus that causes COVID-19—affects different systems in the body, and identify immune correlates. This regulatory science project could potentially help inform development and evaluation of medical countermeasures for COVID-19, according to the FDA.
The $1.55M contract was awarded to Stanford’s Garry P. Nolan, PhD, project leader, and will cover the period of October 2020 through April 2022.
The COVID-19 pandemic—caused by SARS-CoV-2—is the third deadly human outbreak in less than 20 years caused by a zoonotic coronavirus jumping the species barrier. “To help mitigate this public health emergency, the scientific community needs to better understand the pathogenesis of and immune response to SARS-CoV-2 and other coronaviruses. Generating knowledge databases to inform medical countermeasure (MCM) development against SARS-CoV-2 and other coronaviruses is critical to the COVID-19 response, and to preparedness for future outbreaks.
In this Medical Countermeasures Initiative (MCMi) regulatory science project, Stanford University School of Medicine will conduct research to help explain the host factors contributing to coronavirus immune responses, to further the ability to more rapidly predict patient outcomes, and to address unmet needs in patient care.
Researchers will use innovative analytical tools to perform an in-depth analysis of existing tissue samples from previously conducted clinical and nonclinical studies. They will profile circulating immune signatures of coronavirus infection and complete cutting-edge COVID-19 pathology tissue imaging, leveraging novel tools to define the characteristics of tissue viral reservoirs (cell types or areas of the body where the virus persists), and learning more about how SARS-CoV-2 affects different systems in the body.
The project will identify immune correlates of protection, which could potentially help identify and inform development of new coronavirus MCMs, such as drug and vaccine candidates. The project will also help enhance understanding and use of immune correlates for the regulatory review of MCMs.
Collaborators include:
- Public Health England (UK)
- Integrated Research Facility at the National Institutes of Health (NIH)
- California National Primate Research Center at UC Davis
- Erasmus University Medical Center
- Wisconsin National Primate Research Center (WNPRC)
- Harvard Medical School Center for Virology and Vaccine Research
- University of Pittsburgh Graduate School of Public Health
- University of Liverpool (UK)
- University of California San Francisco
- Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán (INCMNSZ) (Mexico)
- Inflammation in COVID-19 – Exploration of Critical Aspects of Pathogenesis (ICECAP) consortium
- University Hospital Basel & Cantonal Hospital of Liestal, Switzerland
During this project, the Stanford University School of Medicine will perform tissue imaging and analysis of samples from existing clinical and nonclinical SARS-CoV-2 studies. The primary outcomes of this project are:
- To conduct multiplexed single-cell analysis of blood samples using CyTOF mass cytometry—a technology that combines flow cytometry and mass spectrometry to simultaneously measure dozens of features located on and in cells—to identify circulating immune cell responses related to stage of disease and severity of outcome following coronavirus infection. Parallel assessments of cases in humans and nonhuman primates (NHPs) will shed light on similarities and differences across species.
- To perform multiplexed antibody-based imaging of respiratory and immune tissues collected from COVID-19 patients or during NHP coronavirus challenge studies being conducted and funded outside of this project, using CO-Detection by indEXing (CODEX). The team will use computational tools to extract single-cell data from CODEX images to precisely identify cell types responding to coronavirus infection, and the spatial relationships of these cells contributing to effective or ineffective immune responses.
- To apply a new technique merging multiplexed antibody-based protein measurements with viral RNA detection to analyze coronavirus infection. This method, called viral Multiplexed Ion Beam Imaging (viralMIBI), will also be applied to tissues from previous clinical and nonclinical studies to gain a deeper understanding of the relationships between presence of viral RNA and viral and host proteins during coronavirus infections.
This project was funded through the MCMi Regulatory Science Extramural Research program.